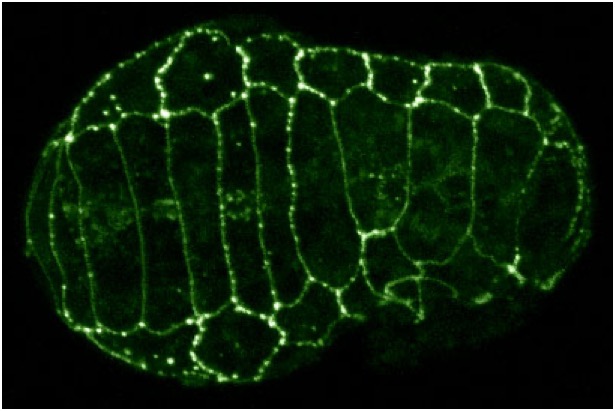
 C. elegans embryo expressing AJM-1::GFP in which the founder cell Cpaa was ablated to remove the left-hand dorsal epidermal cells [Ryan King].
C. elegans embryo expressing AJM-1::GFP in which the founder cell Cpaa was ablated to remove the left-hand dorsal epidermal cells [Ryan King].Dorsal epidermal cells comprise two rows of epithelial cells that lie along the dorsal midline and extend along much of the anterior-posterior axis. These cells intercalate to form a single row of cells. Since there are only 20 cells that intercalate, it is perhaps the simplest known system for studying directed cell rearrangement during embryonic development. Click on the links to see examples videos…
Dorsal intercalation in a wild-type embryo visualized using Nomarski microscopy. Anterior to the left, dorsal view. (30 sec/frame). E. Williams-Masson (Williams-Masson et al., 1998; PubMed).
Download | Play movie
Dorsal intercalation in a wild-type embryo visualized using a dlg-1::gfp translational fusion (Koeppen et al., 2001; PubMed). Frames were acquired at 5 min intervals.
Download | Play movie
We are taking several approaches to study dorsal intercalation that include:
(1) Determining how a Rac/RhoG-dependent actomyosin network regulates intercalation: We developed ways to analyze Rac-dependent motility (Walck-Shannon et al, 2015. Development 142:3549-3560). More recently we extended this network to include the guanine nucleotide exchange factor TIAM-1 (Zhu et al, 2024. J. Cell Sci. 137(5):jcs261509. (*co-first authors) PubMed
(2) Determining what pathways orient cells during dorsal intercalation. We showed that CDC-42 is involved in orienting protrusions as cells begin to migrate (Walck-Shannon et al, 2016. PLOS Genetics 12(11): e1006415). We are now examining other signaling pathways that regulate intercalation.
(3) Determining what transcriptional networks regulate dorsal intercalation: We have returned to classic work from our group (Heid et al, 2001. Dev Biol 236, 165-180) using modern cell lineage tracing, CRISPR/Cas9 genome editing, RNAseq, and other technologies to investigate gene regulatory networks upstream of intercalation.
 THE HARDIN LAB
THE HARDIN LAB