Images in the banner (from left to right and top to bottom): DLG-1/Discs large::GFP during dorsal intercalation [M. Köppen]; Pdlg-1::GFP during ventral enclosure [M. Sheffield]; AJM-1 (green) and muscle (red) during elongation [P. Heid]; HMP-2/beta-catenin electrostatic surface [H.-J. Choi]; AJM-1 (green) and muscle (red) in an elongated embryo [A. Cox-Paulson]; phalloidin staining in elongated embryo [M. Costa]
Interested in working with our team? We're always interested in talking with highly motivated scientists. Click here for more information…
Our work is supported by the National Institute of General Medical Sciences, NIH (current award #R35 GM145312).

CONTACT
Jeff Hardin • Raymond E. Keller and Wayland Noland Distinguished Professor
Department of Integrative Biology, University of Wisconsin-Madison
327 Integrative Biology Research Building • 1117 W. Johnson St. • Madison, WI 53706
voice: (608) 262-9634 • lab: (608) 265-2520 • email: jdhardin@wisc.edu
GRADUATE PROGRAMS
Integrative Biology | Biophysics | Cellular & Molecular Biology | Genetics | Molecular & Cellular Pharmacology
Page last updated: 1/30/26
A new collaboration with Bruce Vogel's group was published in Molecular Biology of the Cell:
Oshima Y, Nagidi Y, Moorthy ME, Heier J, Matsubara JA, Hardin J, Flajnik M, Vogel BE. (2026). TEP-1, a glial thioester protein is required for cilia organization and intraflagellar transport in ensheathed sensory neurons in C. elegans. Mol Biol Cell Jan 28:mbcE25110547. PubMed
Updated the web site to UW Theme 2.0 specifications…
August 2025
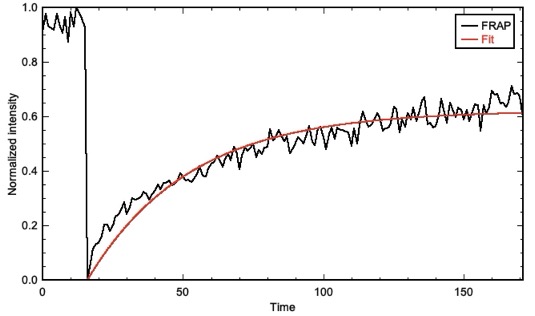 Output from FRAP Analysis, a Jython script for FRAP analysis.
Output from FRAP Analysis, a Jython script for FRAP analysis.
I (Jeff) finally got around to creating and populating two Fiji/ImageJ update sites: 4d-Tools and FRAP-Tools. As the names imply, these contain macros/plugins for 4d microscopy and scripts for analyzing FRAP data, respectively. You can learn more on our Microscopy page.
May 2025
 Kally Arnzen (on the left) defended her Masters thesis on the role of vang-1/Vangl during dorsal intercalation. Way to go, Kally!
Kally Arnzen (on the left) defended her Masters thesis on the role of vang-1/Vangl during dorsal intercalation. Way to go, Kally!
December 2024
 Lilia Thomas (B.S., University. of Illinois), a first-year Biophysics PhD student, joined the lab. Welcome, Lilia!
Lilia Thomas (B.S., University. of Illinois), a first-year Biophysics PhD student, joined the lab. Welcome, Lilia!
September 2024
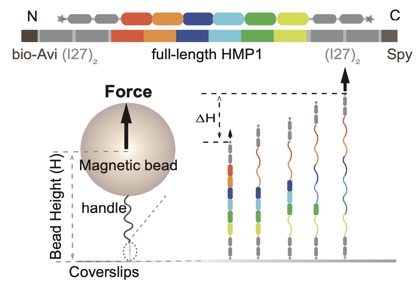
Our collaboration with the Yan lab, Mechanobiology Institute, Singapore is out at PNAS. Le et al (2024). Single-molecule force spectroscopy reveals intra- and intermolecular interactions of C. elegans HMP-1 during mechanotransduction. PNAS 2024 Sep 10;121(37):e2400654121. PubMed
February 2024
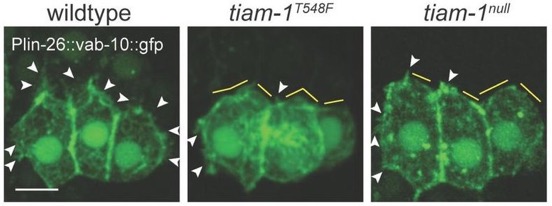
*Zhu, Y., *Tesone, Z., Tan, M., and Hardin, J. (2024). TIAM-1 regulates polarized protrusions during dorsal intercalation in the C. elegans embryo through both its GEF and N-terminal domains. J. Cell Sci. 137(5):jcs261509. (*co-first authors) PubMed
September 2023
Welcome to Adelle Markle (CMB PhD student) and Kally Arnzen (iBio MS student). We're excited to have two new young scientists join us!
August 2023

Yuyun Zhu graduated with a PhD in Genetics. Congratulations, Yuyun!
June 2023
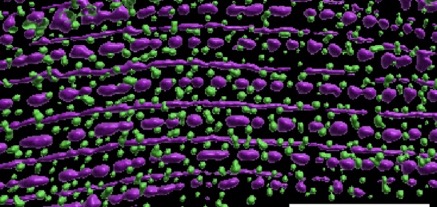
Blake Martin's paper, work done in collaboration with Guy Benian's group, is out in Molecular Biology of the Cell.:
Martin, S.C.T., Qadota, H., Oberhauser, A.F., Hardin, J., and Benian, G.M. (2023). FARL-11 (STRIP1/2) is required for sarcomere and sarcoplasmic reticulum organization in C. elegans. MBoC 34:ar86, 1–13, August 1, 2023. PubMed
February 2023
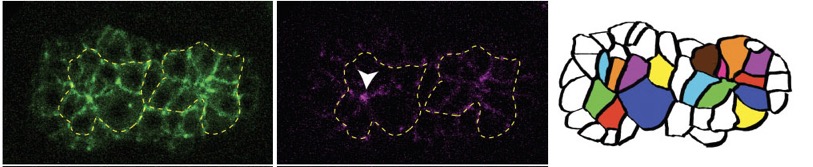
Joel's second paper is out in PLoS Genetics: Serre, J.M. Mark M. Slabodnick, M.S., Goldstein, B. and Hardin, J. (2022). SRGP-1/srGAP and AFD-1/Afadin stabilize HMP-1/α-Catenin at rosettes to seal internalization sites following gastrulation in C. elegans. PLoS Genet. 2023 Mar 3;19(3):e1010507. doi: 10.1371/journal.pgen.1010507. PubMed
September 2022
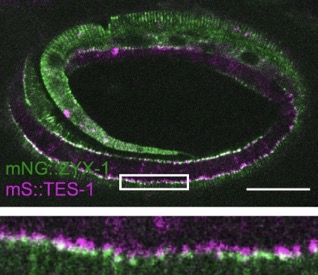
Our paper on TES-1/Tes and ZYX-1/Zyxin is out in Current Biology:
Lynch, A.M.*, Zhu, Y.*, Lucas, B.G., Winkelman, J.D., Bai, K., Martin, S.C.T. , Samuel Block, S., Slabodnick, M.S., Audhya, A., Goldstein, B., Pettitt, J., Gardel, M.L., and Hardin, J.(2022). TES-1/Tes and ZYX-1/Zyxin protect junctional actin networks under tension during epidermal morphogenesis in the C. elegans embryo. Curr. Biol., https://doi.org/10.1016/j.cub.2022.10.045 (*= co-first authors). PubMed
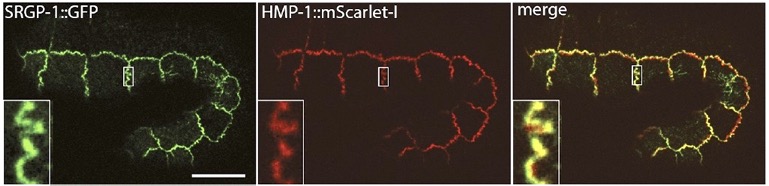
Our paper on SRGP-1/srGAP and the HMP-1/α-catenin M domain is out:
Serre, J.M., Lucas, B., Martin, S.C.M. , Heier, J.A., Xiangqiang Shao, X., and Jeff Hardin, J. (2022). C. elegans srGAP is an α-catenin M domain-binding protein that strengthens cadherin-dependent adhesion during morphogenesis. Development 149 (18): dev200775. . PubMed
July 2022

Joel Serre graduated with a PhD in Genetics. Way to go, Joel!
May 2022

Blake Martin graduated with a PhD in Biophysics. He's headed to the Özpolat group at Washington Univ, in St. Louis. Congratulations, Blake!

Blake Martin and former postdoc Joanna Bundus, with their colleagues (including Blake's family!) have made major strides in creating a Diné (Navajo) lexicon through Project ENABLE (Enriching Navajo as a Language for Biology Education).
February 2022
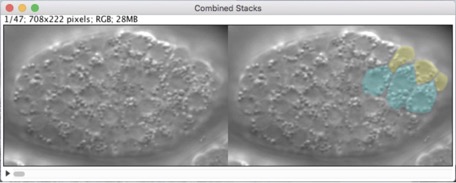 Our new methods chapter is out!
Our new methods chapter is out!
Hardin, J., Serre, J., King, R., Walck-Shannon, E., and Reiner, D. (2022). Imaging epidermal cell rearrangement in the C. elegans embryo. Methods Mol Biol 2438:345-376. PubMed
January 2022
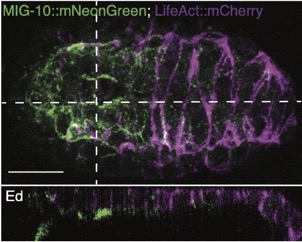 Joel's work on mig-10/lamellipodin is out as a microPublication:
Joel's work on mig-10/lamellipodin is out as a microPublication:
Serre, JM; Hardin, J (2022). The Lamellipodin homologue MIG-10 is not essential for dorsal intercalation in the embryonic epidermis of the C. elegans embryo. microPublication Biology. https://doi.org/10.17912/micropub.biology.000522 PubMed
Welcome to the Hardin Lab
We use the C. elegans embryo as a model for investigating cell movement and cell adhesion during embryonic development. Understanding how cells move, and how they make and break adhesions has important implications for understanding birth defects during human development and for understanding cancer progression. To find out more, click here...Interested in working with our team? We're always interested in talking with highly motivated scientists. Click here for more information…
Our work is supported by the National Institute of General Medical Sciences, NIH (current award #R35 GM145312).

CONTACT
Jeff Hardin • Raymond E. Keller and Wayland Noland Distinguished Professor
Department of Integrative Biology, University of Wisconsin-Madison
327 Integrative Biology Research Building • 1117 W. Johnson St. • Madison, WI 53706
voice: (608) 262-9634 • lab: (608) 265-2520 • email: jdhardin@wisc.edu
GRADUATE PROGRAMS
Integrative Biology | Biophysics | Cellular & Molecular Biology | Genetics | Molecular & Cellular Pharmacology
Page last updated: 1/30/26
Recent News
January 2026A new collaboration with Bruce Vogel's group was published in Molecular Biology of the Cell:
Oshima Y, Nagidi Y, Moorthy ME, Heier J, Matsubara JA, Hardin J, Flajnik M, Vogel BE. (2026). TEP-1, a glial thioester protein is required for cilia organization and intraflagellar transport in ensheathed sensory neurons in C. elegans. Mol Biol Cell Jan 28:mbcE25110547. PubMed
Updated the web site to UW Theme 2.0 specifications…
August 2025
 Output from FRAP Analysis, a Jython script for FRAP analysis.
Output from FRAP Analysis, a Jython script for FRAP analysis.I (Jeff) finally got around to creating and populating two Fiji/ImageJ update sites: 4d-Tools and FRAP-Tools. As the names imply, these contain macros/plugins for 4d microscopy and scripts for analyzing FRAP data, respectively. You can learn more on our Microscopy page.
May 2025
 Kally Arnzen (on the left) defended her Masters thesis on the role of vang-1/Vangl during dorsal intercalation. Way to go, Kally!
Kally Arnzen (on the left) defended her Masters thesis on the role of vang-1/Vangl during dorsal intercalation. Way to go, Kally!December 2024
 Lilia Thomas (B.S., University. of Illinois), a first-year Biophysics PhD student, joined the lab. Welcome, Lilia!
Lilia Thomas (B.S., University. of Illinois), a first-year Biophysics PhD student, joined the lab. Welcome, Lilia!September 2024

Our collaboration with the Yan lab, Mechanobiology Institute, Singapore is out at PNAS. Le et al (2024). Single-molecule force spectroscopy reveals intra- and intermolecular interactions of C. elegans HMP-1 during mechanotransduction. PNAS 2024 Sep 10;121(37):e2400654121. PubMed
February 2024

*Zhu, Y., *Tesone, Z., Tan, M., and Hardin, J. (2024). TIAM-1 regulates polarized protrusions during dorsal intercalation in the C. elegans embryo through both its GEF and N-terminal domains. J. Cell Sci. 137(5):jcs261509. (*co-first authors) PubMed
September 2023
Welcome to Adelle Markle (CMB PhD student) and Kally Arnzen (iBio MS student). We're excited to have two new young scientists join us!
August 2023

Yuyun Zhu graduated with a PhD in Genetics. Congratulations, Yuyun!
June 2023

Blake Martin's paper, work done in collaboration with Guy Benian's group, is out in Molecular Biology of the Cell.:
Martin, S.C.T., Qadota, H., Oberhauser, A.F., Hardin, J., and Benian, G.M. (2023). FARL-11 (STRIP1/2) is required for sarcomere and sarcoplasmic reticulum organization in C. elegans. MBoC 34:ar86, 1–13, August 1, 2023. PubMed
February 2023

Joel's second paper is out in PLoS Genetics: Serre, J.M. Mark M. Slabodnick, M.S., Goldstein, B. and Hardin, J. (2022). SRGP-1/srGAP and AFD-1/Afadin stabilize HMP-1/α-Catenin at rosettes to seal internalization sites following gastrulation in C. elegans. PLoS Genet. 2023 Mar 3;19(3):e1010507. doi: 10.1371/journal.pgen.1010507. PubMed
September 2022

Our paper on TES-1/Tes and ZYX-1/Zyxin is out in Current Biology:
Lynch, A.M.*, Zhu, Y.*, Lucas, B.G., Winkelman, J.D., Bai, K., Martin, S.C.T. , Samuel Block, S., Slabodnick, M.S., Audhya, A., Goldstein, B., Pettitt, J., Gardel, M.L., and Hardin, J.(2022). TES-1/Tes and ZYX-1/Zyxin protect junctional actin networks under tension during epidermal morphogenesis in the C. elegans embryo. Curr. Biol., https://doi.org/10.1016/j.cub.2022.10.045 (*= co-first authors). PubMed

Our paper on SRGP-1/srGAP and the HMP-1/α-catenin M domain is out:
Serre, J.M., Lucas, B., Martin, S.C.M. , Heier, J.A., Xiangqiang Shao, X., and Jeff Hardin, J. (2022). C. elegans srGAP is an α-catenin M domain-binding protein that strengthens cadherin-dependent adhesion during morphogenesis. Development 149 (18): dev200775. . PubMed
July 2022

Joel Serre graduated with a PhD in Genetics. Way to go, Joel!
May 2022

Blake Martin graduated with a PhD in Biophysics. He's headed to the Özpolat group at Washington Univ, in St. Louis. Congratulations, Blake!

Blake Martin and former postdoc Joanna Bundus, with their colleagues (including Blake's family!) have made major strides in creating a Diné (Navajo) lexicon through Project ENABLE (Enriching Navajo as a Language for Biology Education).
February 2022
 Our new methods chapter is out!
Our new methods chapter is out! Hardin, J., Serre, J., King, R., Walck-Shannon, E., and Reiner, D. (2022). Imaging epidermal cell rearrangement in the C. elegans embryo. Methods Mol Biol 2438:345-376. PubMed
January 2022
 Joel's work on mig-10/lamellipodin is out as a microPublication:
Joel's work on mig-10/lamellipodin is out as a microPublication:Serre, JM; Hardin, J (2022). The Lamellipodin homologue MIG-10 is not essential for dorsal intercalation in the embryonic epidermis of the C. elegans embryo. microPublication Biology. https://doi.org/10.17912/micropub.biology.000522 PubMed
Older News
November 2021We've finally moved to the modern era! Please update your bookmarks from http://worms.zoology.wisc.edu to https://worms.zoology.wisc.edu
Thanks to Jon Hardin, CEO of Hardin Design and Development, for helping his dad upgrade the site.
October 2021
Welcome to Zoe Tesone (new CMB grad student) and Jon Heier (new postdoc, who joins us from Adam Kwiatkowski's lab at Pitt)! Congrats to former postdoc Joanna Bundus on becoming Vice President for Research at Xylome Corporation.
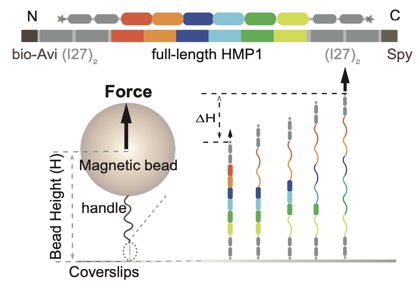
A preprint on our collaboration with the Yan lab, Mechanobiology Institute, Singapore is now available to preview on bioRxiv: Le et al (2021). Mechano-Biochemical Regulation of the C. elegans HMP1–HMP2 protein complex. bioRxiv 2021.09.27.462086; doi: https://doi.org/10.1101/2021.09.27.462086. Link
March 2021
I (Jeff) am pleased to say that World of the Cell 10th edition has been released! Many thanks to my coauthor, James Lodolce and the Pearson team for a job well done.
August 2020
The movies on this site have finally been upgraded to the modern HTML5 era! Now they can all be viewed on smart phones and tablets without QuickTime.
Hardin, J. and Weliky, M. (2019). Cell rearrangement induced by filopodial tension accounts for the late phase of convergent extension in the sea urchin archenteron. Mol. Biol. Cell 30:1911-1919. PubMed
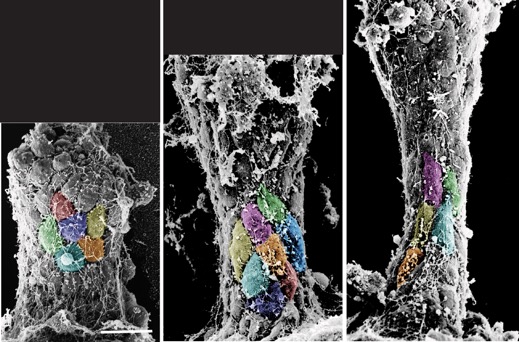 Colorzed SEMs of L. pictus gastrulae (J. Hardin).
Colorzed SEMs of L. pictus gastrulae (J. Hardin).Shao, X., Lucas, B.G., Strauch, J. and Hardin, J. (2019). The adhesion modulation domain of C. elegans α-catenin regulates actin binding during morphogenesis. Mol. Biol. Cell 30:2115-2123. PubMed
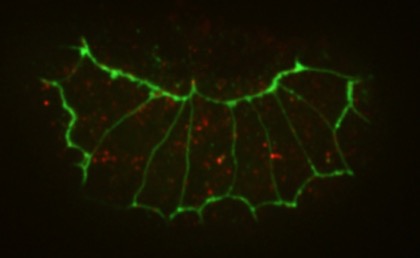 aanti-AJM-1 (green) and HMP-1 (red) staining in a hmp-1(jc48) (a CRISPR null allele; Xiangqiang Shao).
aanti-AJM-1 (green) and HMP-1 (red) staining in a hmp-1(jc48) (a CRISPR null allele; Xiangqiang Shao).November 2017
Tim's paper got the cover of MBoC for December 2017!
Loveless, T., Qadota, H., Benian, GM., and Hardin, J. (2017). C. elegans SORB-1 localizes to integrin adhesion sites and is required for organization of sarcomeres and mitochondria in myocytes. Mol. Bio. Cell, 28:3621-3633. PubMed
October 2017
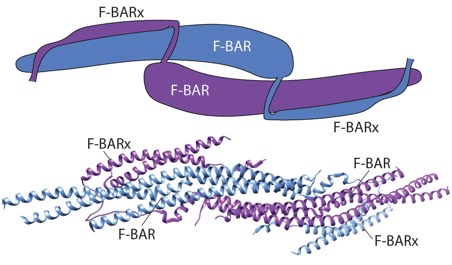
srGAP review: Lucas, B. and Hardin, J. (2017). Mind the (sr)GAP – roles of Slit-Robo GAPs in neurons, brains and beyond. J. Cell Sci. 130: 3965-3974. PubMed
August 2017
Fine-mapping α-/β-catenin binding interface in vitro and in vivo (with Hee-Jung Choi's group). Shao, X. Kang, H., Loveless, T., Lee, G.R., , Seok, C., Weis, W.I., and Choi, H.-J., and Hardin, J. (2017). Cell–cell adhesion in metazoans relies on evolutionarily conserved features of the α-catenin•β-catenin–binding interface. J. Biol. Chem. 292,16477–16490. PubMed
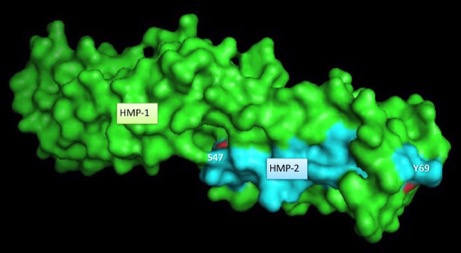 Surface rendering of HMP-1/α-catenin (green) and HMP-2/β-catenin (aqua). Rendering by Tim Loveless
Surface rendering of HMP-1/α-catenin (green) and HMP-2/β-catenin (aqua). Rendering by Tim LovelessJune 2017
Student news: Congratulations to new Genetics PhDs Drs. Bethany Lucas and Xiangqiang Shao, who both defended their theses.
May 2017
The structure of a C. elegans α-catenin. Kang et al (2017). Structural and functional characterization of Caenorhabditis elegans α-catenin reveals constitutive binding to β-catenin and actin. (with Hee-Jung Choi's group; J. Biol. Chem. 29, 7077-7086). PubMed
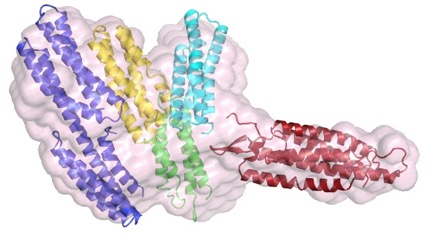 The N terminus (purple, yellow), M domain (green, aqua) and a homology model of the actin-binding domain (red) of HMP-1/α-catenin fitted into the SAXS envelope of HMP-1. Rendering by Hee-Jung Choi.November 2016
The N terminus (purple, yellow), M domain (green, aqua) and a homology model of the actin-binding domain (red) of HMP-1/α-catenin fitted into the SAXS envelope of HMP-1. Rendering by Hee-Jung Choi.November 2016Walck-Shannon et al. (2016). CDC-42 orients cell migration during epithelial intercalation in the Caenorhabditis elegans epidermis. PLOS Genetics 12(11): e1006415 PubMed
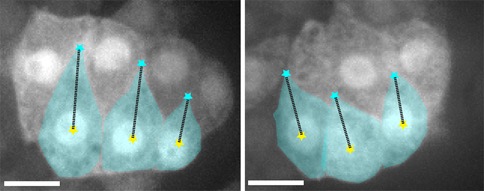 Confocal images of an epidermal cytoplasmic reporter (Plbp-1::GFP) in wild-type (WT) (left) and ZF1::cdc-42; cdc-42(gk388) (right) embryos, in which there is late maternal loss of CDC-42, 45 min. after terminal division. Scale bar is 5 μm. [Elise Walck-Shannon]
Confocal images of an epidermal cytoplasmic reporter (Plbp-1::GFP) in wild-type (WT) (left) and ZF1::cdc-42; cdc-42(gk388) (right) embryos, in which there is late maternal loss of CDC-42, 45 min. after terminal division. Scale bar is 5 μm. [Elise Walck-Shannon]February 2016
F1000 review of the cadherin/catenin complex in C. elegans. F1000Research 2015, 4(F1000 Faculty Rev):1473 (doi: 10.12688/f1000research.6866.1) PubMed
World of the Cell 9th edition (Pearson) is out! Thanks to Marisa Otegui for the beautiful image used on the cover.
October 2015
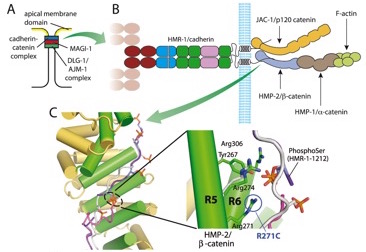
Proteomic analysis of the cadherin/catenin complex in C. elegans (collaboration with Jon Audhys's group):
Callaci, S., Morrison, K., Shao, X., Schuh, A.L. Wang, Y., Yates III, J.R., Hardin, J., and Audhya, A. (2015). Phosphoregulation of the C. elegans cadherin-catenin complex. Biochemistry 472:339-52.. PubMed
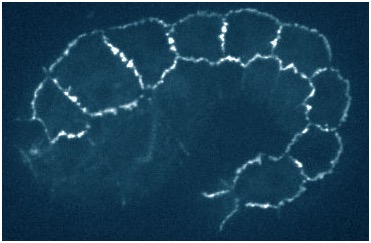 hmp-1 mutant embryo rescued with a quadruple mutant phosphomimetic form of HMP-1 tagged with gfp (Xiangqiang Shao).
hmp-1 mutant embryo rescued with a quadruple mutant phosphomimetic form of HMP-1 tagged with gfp (Xiangqiang Shao).August 2015
Dorsal intercalation in C. elegans uses a conserved Trio/CARMIL system upstream of Rac and RhoG. Development 142, 3549-3560. PubMed
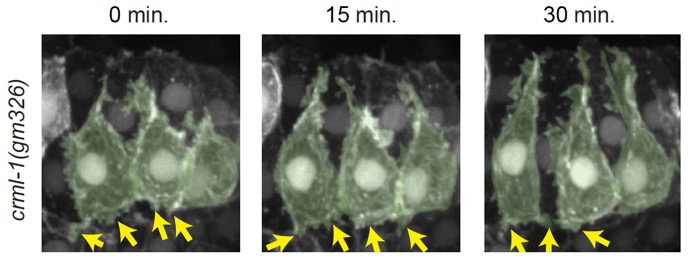 crml-1(gm326) dorsal cells have excessive protrusive activity, which can be suppressed by GEF1 loss of function n unc-73(rh40) mutants. Left-hand cells are psuedocolored green. Yellow arrows denote excessive, lateral protrusions. Scale bar is 5 μm. [Elise Walck-Shannon]
crml-1(gm326) dorsal cells have excessive protrusive activity, which can be suppressed by GEF1 loss of function n unc-73(rh40) mutants. Left-hand cells are psuedocolored green. Yellow arrows denote excessive, lateral protrusions. Scale bar is 5 μm. [Elise Walck-Shannon]July 2015
Student news: Congratulations to Elise Walck-Shannon, who defended her thesis July 29, 2015!
Blake Martin received a Gilliam fellowship from HHMI!
March 2015
Angstroms to embryos: structure-function analysis of the cadherin complex in C. elegans
(collaboration with Bill Weis, Stanford, and Hee-Jung Choi, Seoul Nat. Univ.; Developmental Cell 33, 82–93)
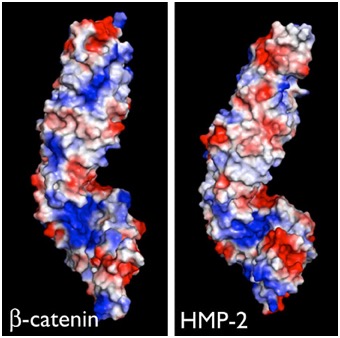 Electrostatic representation of zebrafish β-catenin (left) and C. elegans HMP-2 (right). [Hee-Jung Choi, Bill Weis, Tim Loveless]
Electrostatic representation of zebrafish β-catenin (left) and C. elegans HMP-2 (right). [Hee-Jung Choi, Bill Weis, Tim Loveless]June 2014
 | Minor updates to ImageJ plugins are available. Go to the Microscopy page. |
January 2014
Cell rearrangement review
Walck-Shannon, E. and Hardin, J. (2014). Cell intercalation from top to bottom. Nature Rev. Mol. Cell. Bio 15:34-48. Pubmed
February 2013
Mapping functionally important domains in the C terminus of α-catenin
Maiden, S.L., Harrison, N., Keegan, J., Cain, B., Lynch, A.M., Pettitt, J., and Hardin, J. Specific conserved C-terminal amino acids of Caenorhabditis elegans HMP-1/α-catenin modulate F-actin binding independently of vinculin. J. Biol. Chem. 288:5694-706. PubMed
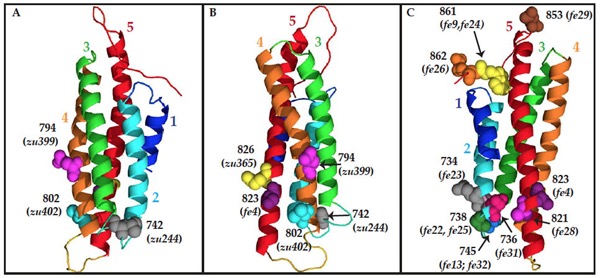 Homology model of the C terminus o HMP-1/α-catenin, using the metavinculin VH3 domain crystal structure as a template. Residues mutated in hmp-1 alleles are shown as space-filled molecules labeled with the amino acid number. (A) and (B) Residues mutated in strong loss-of-function alleles. (C) Residues mutated in hmp-1(fe4) and intragenic suppressor alleles. [Stephanie Maiden]
Homology model of the C terminus o HMP-1/α-catenin, using the metavinculin VH3 domain crystal structure as a template. Residues mutated in hmp-1 alleles are shown as space-filled molecules labeled with the amino acid number. (A) and (B) Residues mutated in strong loss-of-function alleles. (C) Residues mutated in hmp-1(fe4) and intragenic suppressor alleles. [Stephanie Maiden]August 2012
MAGI-1, AFD-1/afadin, and the cadherin interactome during morphogenesis
Lynch, A.M., Grana, T., Cox-Paulson, E., Couthier, A., Cameron, M., Chin-Sang, I., Pettitt, J., and Hardin, J. (2012). A genome-wide functional screen identifies MAGI-1 as an L1CAM-dependent stabilizer of apical junctions in C. elegans. Curr. Biol 22, 1891–1899. PubMed
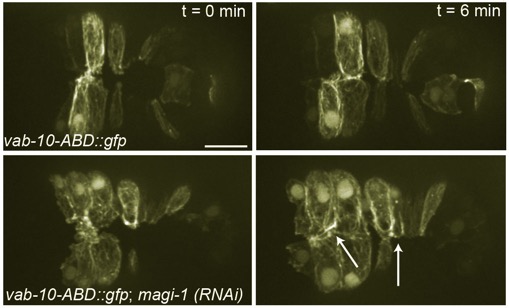 Top: Wild-type embryo expressing an actin reporter during ventral enclosure. Bottom: a magi-1(RNAi) embryo. Cells migrate ventrally, but migration is irregular and cells display excess protrusive activity at the ventral midline. [Allison Lynch]
Top: Wild-type embryo expressing an actin reporter during ventral enclosure. Bottom: a magi-1(RNAi) embryo. Cells migrate ventrally, but migration is irregular and cells display excess protrusive activity at the ventral midline. [Allison Lynch]June 2012
Tropomodulin protects adherens junctions under stress during morphogenesis
Cox-Paulson, E., Walck-Shannon, E., Lynch, A., Yamashiro, S., Zaidel-Bar, R., Celeste C. Eno, C., Ono, S., and Hardin, J. (2012). Tropomodulin protects α-catenin-dependent junctional actin networks under stress during epithelial morphogenesis. Curr. Biol. 22:1500-1505. PubMed
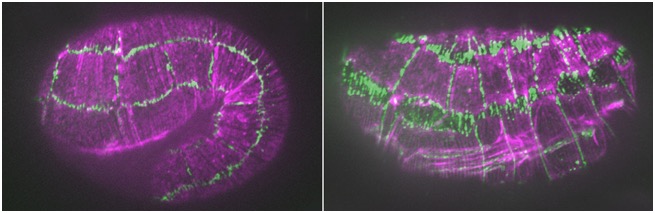 hmp-1/α-catenin and unc-94/tropomodulin synergistically regulate embryonic morphogenesis. Purple, F-actin (phalloidin staining); green, JAC-1/p120ctn::GFP. Left: a hmp-1(fe4) embryo at the 1.5-fold stage. Junctions are largely normal. Right: a hmp-1(fe4);unc-94(RNAi) embryo. Junctions between seam cells and ventral and dorsal epidermal cells have ripped apart [Abbi Cox-Paulson]
hmp-1/α-catenin and unc-94/tropomodulin synergistically regulate embryonic morphogenesis. Purple, F-actin (phalloidin staining); green, JAC-1/p120ctn::GFP. Left: a hmp-1(fe4) embryo at the 1.5-fold stage. Junctions are largely normal. Right: a hmp-1(fe4);unc-94(RNAi) embryo. Junctions between seam cells and ventral and dorsal epidermal cells have ripped apart [Abbi Cox-Paulson]January 2012
Cell migration during ventral enclosure
Ikegami, R., Simokat, K., Zheng, H., Dixon, L., Garriga, G., Hardin, J. and Culotti, J. (2012). Semaphorin and Eph receptor signaling guide a series of cell movements for ventral enclosure in C. elegans. Curr. Biol. 22:1–11. PubMed
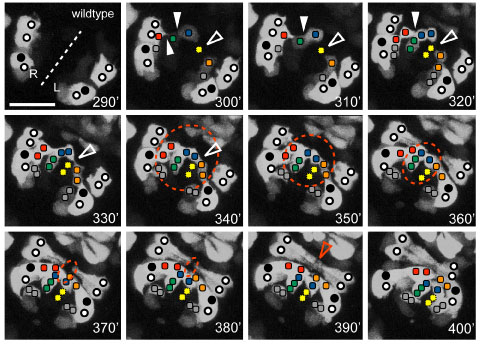 Ventral view of the pocket region of an embryo expressing Pplx-2::gfp; anterior at top right. The ventral midline (dashed line) and relative positions of expressing identified P cells (circles, closed circles are P9/10) and plexin expressing cells on the surface of the open ventral pocket (colored squares) are indicated. [Kristin Simokat and Richard Ikegami]
Ventral view of the pocket region of an embryo expressing Pplx-2::gfp; anterior at top right. The ventral midline (dashed line) and relative positions of expressing identified P cells (circles, closed circles are P9/10) and plexin expressing cells on the surface of the open ventral pocket (colored squares) are indicated. [Kristin Simokat and Richard Ikegami]November 2011
α-catenin review
Maiden, S.L. and Hardin, J. (2011). The secret life of α-catenin: moonlighting during morphogenesis. J. Cell Biol. 195:543–552. PubMed
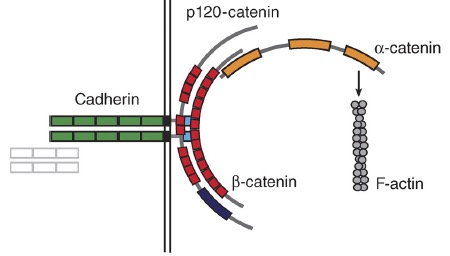
January 2011
Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells
Neukomm, L.J., Frei, A.P., Cabello, J., Kinchen, J.M., Zaidel-Bar, R., Ma, Z., Haney, L.B., Hardin, J., Ravichandran, K.S., Moreno, S., and Hengartner, M.O. (2011). Loss of the RhoGAP SRGP1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nature Cell Biol. 13:79-86. PubMed
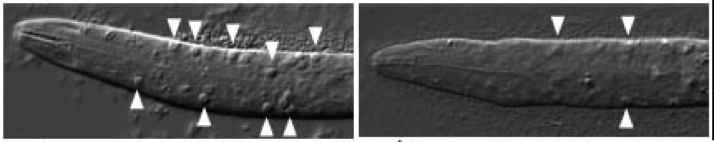
ced-6 larvae accumulate cell corpses due to engulfment defects (arrowheads, left). Engulfment defects are suppressed in ced-6;srgp-1 double mutants (right). [Lukas Neukomm]
November 2010
SRGP-1/srGAP regulates membrane protrusion during cell-cell adhesion
(Zaidel-Bar, R., Joyce, M.J., Lynch, A.M., Witte, K., Audhya, A., and Hardin, J. (2010). The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J. Cell Biol. 191, 761-9.) PubMed JCB In This Issue
 November 15, 2010 cover! November 15, 2010 cover! | 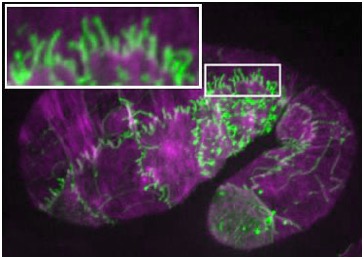 Embryo expressing SRGP-1::GFP (green) stained for actin (purple).The inset shows extensive induced tubulations. [Ronen Zaidel-Bar] Embryo expressing SRGP-1::GFP (green) stained for actin (purple).The inset shows extensive induced tubulations. [Ronen Zaidel-Bar] |
August 2010
Intramolecular regulation of alpha-catenin's ability to bind actin
(Kwiatkowski, A.V., Maiden, S.L., Pokutta, S., Choi, H.-J., Benjamin, J.M., Lynch, A.M., Nelson, W.J., Weis, W.I., and Hardin, J. (2010). In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. PNAS 107,14591-14596.)
PubMed
 August 17, 2010 cover! August 17, 2010 cover! | <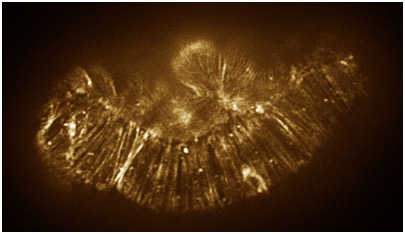 Color adjusted image of a hmp-1(zu278) mutant embryo stained with phalloidin. [Stephanie Maiden]. Color adjusted image of a hmp-1(zu278) mutant embryo stained with phalloidin. [Stephanie Maiden]. |
May 2010
Cadherins and L1CAMs during cell-cell adhesion and gastrulation
(Grana, T.M., Cox, E.A., Lynch, A.M., and Hardin, J. (2010). SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation. Dev. Biol. 344, 731–744.)
PubMed
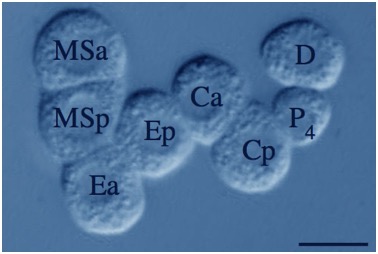 Color adjusted image of a devitellinized hmr-1(RNAi);sax-7(eq1) C. elegans embryo. Blastomeres are loosely adherent and cell division orientations are abnormal. [Theresa Grana].
Color adjusted image of a devitellinized hmr-1(RNAi);sax-7(eq1) C. elegans embryo. Blastomeres are loosely adherent and cell division orientations are abnormal. [Theresa Grana]. The Hardin Lab
The Hardin Lab