The F-BAR domain of SRGP-1 facilitates cell–cell adhesion during C. elegans morphogenesis
Ronen Zaidel-Bar,1 Michael J. Joyce,1 Allison M. Lynch,2 Kristen Witte,3 Anjon Audhya,3 and Jeff Hardin1,2
1Department of Zoology, 2Graduate Program in Genetics, and 3Department of Biomolecular Chemistry, University of Wisconsin–Madison, Madison, WI 53706
Supplemental material for the following paper:
Zaidel-Bar, R., Joyce, M.J., Lynch, A.M., Witte, K., Audhya, A., and Hardin, J. (2010). The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J. Cell Biol. 191, 761-9. PubMed
Summary
Supplemental Figure 1
Supplemental Figure 2
Supplemental Figure 3
Video sequence 1
Video sequence 2
Video sequence 3
Video sequence 4
Summary
Robust cell–cell adhesion is critical for tissue integrity and morphogenesis, yet little is known about the molecular mechanisms controlling cell–cell junction architecture and strength. We discovered that SRGP-1 is a novel component of cell–cell junctions in Caenorhabditis elegans, localizing via its F-BAR (Bin1, Amphiphysin, and RVS167) domain and a flanking 200–amino acid sequence. SRGP-1 activity promotes an increase in membrane dynamics at nascent cell–cell contacts and the rapid formation of new junctions; in addition, srgp-1 loss of function is lethal in embryos with compromised cadherin–catenin complexes. Conversely, excess SRGP-1 activity leads to outward bending and projections of junctions. The C-terminal half of SRGP-1 interacts with the N-terminal F-BAR domain and negatively regulates its activity. Significantly, in vivo structure–function analysis establishes a role for the F-BAR domain in promoting rapid and robust cell adhesion during embryonic closure events, independent of the Rho guanosine triphosphatase–activating protein domain. These studies establish a new role for this conserved protein family in modulating cell–cell adhesion.
Return to top
Supplemental Figures

Figure S1. Multiple alignment of C. elegans SRGP-1 with human srGAP family members. The full protein sequences of C. elegans SRGP-1 and human srGAP1, srGAP2, srGAP3, and ARHGAP4 were aligned using ClustalW. A high degree of conservation is found along the F-BAR and GAP domains, as well as regions between them (putative junctional targeting sequence) and immediately after the GAP domain. However, the SH3 domain is not conserved in SRGP-1. Dark blue highlight marks residues that are conserved between all five sequences, and light blue highlight marks residues that are conserved in four out of five of the sequences.
Supplemental Figure 1 - JPEG (1.7 Mb) Download
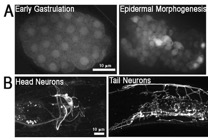
Figure S2. Expression pattern of the srgp-1 promoter. An extrachromosomal array, sEx10607, was used to visualize the expression of cytoplasmic GFP driven by the srgp-1 promoter. (A) In the embryo, srgp-1 is first expressed in all cells, and later, expression is restricted to neuroblasts, epidermal, and pharyngeal cells. (B) In adults, expression is seen in the nervous system as well as in the pharynx and spermatheca.
Supplemental Figure 2 - JPEG (0.3 Mb) Download
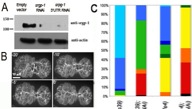
Figure S3. Knockdown of srgp-1 enhances lethality and leads to more severe defects in hmp-1/alpha-catenin and hmp-2/beta-catenin hypomorphs. (A) Western blot showing the knockdown of SRGP-1 by feeding srgp-1(RNAi). Numbers on the left are in given kilodaltons. (B) Quantification of ventral closure rate in control and srgp-1(RNAi) embryos expressing HMP-1::GFP. d is the distance between two opposing epidermal cells that was measured and used to quantify the rate of ventral enclosure as shown in the graph. (C) Nomarski videos of single and double mutants were analyzed to determine the earliest failure and lethal phenotype (defined in the legend). The bar chart represents pooled data from three or more videos for each genotype (n > 60). (D) Comparison of cleft closure in wild-type, hmp-2(qm39), and hmp-2(qm39);srgp-1(RNAi) embryos shows rapid and irreversible sealing of the cleft in wild type compared with adhesion and deadhesion events in the mutants (arrows), which lead to slower closure in the single mutant and widening of the cleft in the double mutant. WT, wildtype. (E) Quantification of cleft area versus time. Lines depict the mean from more than six individual embryos, and error bars denote SEM.
Supplemental Figure 3 - JPEG (0.6 Mb) Download
Return to top
Videos
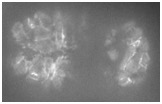
Video 1 (0.8 Mb). Ventral enclosure in wild-type and srgp-1(RNAi) embryos. HMP-1::GFP dynamics show extensive membrane ruffling (arrows) and rapid junction formation in wild type versus less membrane ruffling and slower junction formation in the srgp- 1(RNAi) embryo. Single focal plane. Frames are shown at 1 min apart. Play rate is nine frames per second.
Download
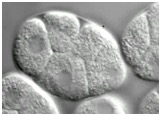
Video 2 (1.8 Mb). Morphogenetic failure in hmp-1(fe4)/alpha-catenin;srgp-1 double mutant embryos. One Z section from a Nomarski time-lapse video following the development of hmp-1(fe4);srgp-1(RNAi) embryos from the four-cell stage. All embryos fail in gastrulation cleft closure and subsequently rupture from the cleft. Play rate is nine frames per second.
Download
Return to top
Video 3 (0.6 Mb). Gastrulation cleft closure in wildtype, hmp-2(qm39)/beta-catenin embryo, and hmp-2(qm39)/beta-catenin;srgp-1(RNAi) embryo. Single Z section, 1-min interval time-lapse videos of cleft closure in wild-type (left), hmp-2(qm39) (middle), and hmp- 2(qm39);srgp-1(RNAi) embryos. Note the faster closure in wild-type embryos and the separation of cells that have already made contact in the mutant embryos. Play rate is nine frames per second.
Download
Video 4 (4.1 Mb). Dynamics of SRGP-1::GFP at various developmental stages. Time-lapse videos of SRGP-1::GFP were acquired at 1- min intervals. Each frame is a projection of eight Z sections spaced 0.2 μm apart. On the top left is a dorsal view of epidermal cells after intercalation; on the top right is a ventral view during late gastrulation. On the bottom left is a view of ventral epidermal enclosure, and on the bottom right is epidermal elongation. Play rate is nine frames per second.
Download
Return to top
 THE HARDIN LAB
THE HARDIN LAB